Accessing the Neuroimaging Data Releases¶
The neuroimaging data releases contain the MRI data, physiological data obtained during scanning (cardiac and respiratory) and only basic phenotypic information (age, sex, handedness). The following scans are included for subjects in the Cross-Sectional Lifespan Connectomics Study, Longitudinal Developmental Connectomics Study, and Mapping Interindividual Variation In The Aging Connectome studies:
- Anatomical (MPRAGE)
- Diffusion Tensor Imaging (137 Directions)
- Resting Functional (TR = 645ms, Multiband)
- Resting Functional (TR = 1400ms, Multiband)
- Resting Functional (TR = 2500ms)
- Visual Checkerboard (TR = 645ms, Multiband)
- Visual Checkerboard (TR = 1400ms, Multiband)
- Breath Hold (TR = 1400ms, Multiband)
The Real-Time Neurofeedback study includes these scans:
- Anatomical (MPRAGE)
- Default Mode Network Training Scan
- Default Mode Network Testing Scan
- Moral Dilemma
- MSIT
- PEER1
- PEER2
Note: For a list of all possible study codes, see here.
Downloading Neuroimaging Data Release Data from the FCP-INDI S3 Bucket¶
Data for the Rockland Sample Neuroimaging Data Release are available for download in an Amazon Web Services S3 bucket. The bucket is publicly available and the path is s3://fcp-indi/data/Projects/RocklandSample/RawDataBIDSLatest. The data is currently organized as follows:
- RawDataBidsLatest : The latest data release of raw data organized in the BIDS format. This folder includes data from all the releases. Since BIDS makes provisions for phenotypic and data collected during scanning (physiological,event-related), this data is also included in this folder in addition to the MRI series NifTIs. DICOMs are not included. AWS Links
- Step-by-step instructions to download: Here you can see here a pdf with step-by-step instruction on how to download neuroimaging data using a bash or a python script.
- RawData : Raw data for Rockland sample releases converted to NifTI format and not separated by release. The directory tree is organized with participants at the root, visits below participants, and individual series below visits.
- RawDataTars : Raw data compressed with Gzip and separated by release.
- RawDataBIDS : Raw data organized in the BIDS format. Since BIDS makes provisions for phenotypic and data collected during scanning (physiological,event-related), this data is also included in this folder in addition to the MRI series NifTIs. DICOMs are not included.
Old Data Releases . We do not recommend downloading data from these folders since they have not been updated and are organized with old study codes:
Each file in the S3 bucket can only be accessed using HTTP (i.e., no ftp or scp ). You can obtain a URL for each desired file here or from LORIS and then download it using the AWS command line interface or an HTTP client such as a web browser, wget, or curl. Each file can only be accessed using its literal name- wildcards will not work. We have written a script (described below) that can perform custom dataset downloads for you via HTTP.
There are file transfer programs that can handle S3 natively and will allow you to navigate through the data using a file browser. Cyberduck is one such program that works with Windows and Mac OS X. Cyberduck also has a command line version that works with Windows, Mac OS X, and Linux.
Instructions for using Cyberduck are as follows:
- Open Cyberduck and click on Open Connection.
- Set the application protocol in the dropdown menu to FTP (File Transfer Protocol).
- Check the Anonymous Login checkbox.
- CHANGE the application protocol in the dropdown menu to Amazon S3.
- SET the server to https://s3.amazonaws.com/fcp-indi/data/Projects/RocklandSample
- CHANGE the application protocol in the dropdown menu to Amazon S3.
- Click Connect.
Due to some issues of connecting to S3 bucket on Cyberduck using an anonymous login, you will need to take the following steps for a workaround:
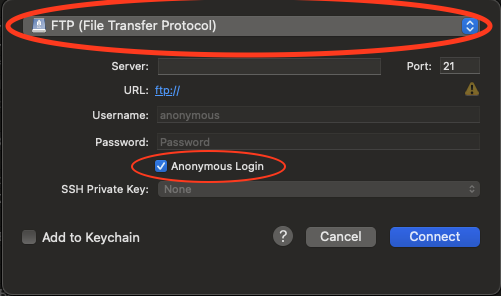
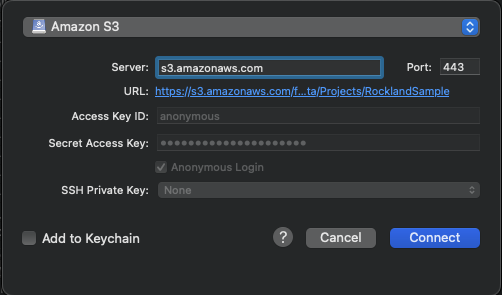
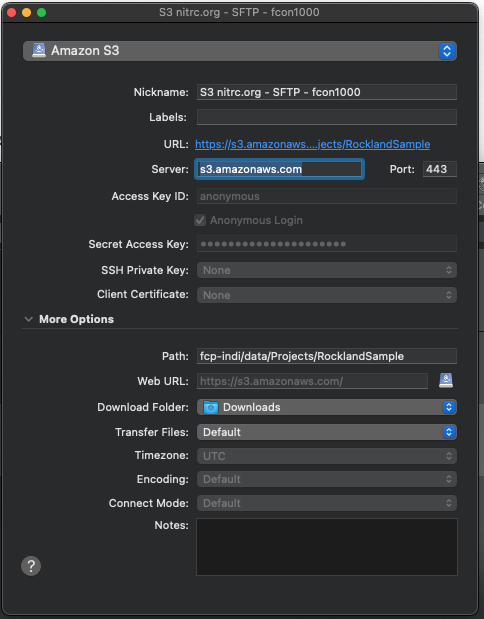
To set Cyberduck Bookmark for Rockland Sample, Instructions are as follows:
- Open Cyberduck and click on Action.
- At the bottom of the dropdown window, click on New Bookmark.
- Set the application protocol in the dropdown menu to FTP (File Transfer Protocol).
- Check the Anonymous Login checkbox.
- CHANGE the application protocol in the dropdown menu to Amazon S3.
- Change the Nickname to Rockland Sample Neuroimaging Amazon S3.
- Make sure the application protocol in the dropdown menu is Amazon S3.
- Make sure Server is set to s3.amazonaws.com .
- Expand the More Options tab and SET Path to fcp-indi/data/Projects/RocklandSample
- Press Enter on your keyboard and the bookmark is saved
- Exit the new bookmark window and the new Rockland Sample bookmark should appear in the bookmark tab
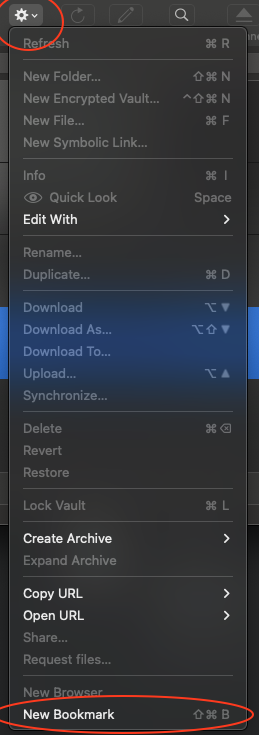
Due to some issues of connecting to S3 bucket on Cyberduck using an anonymous login, you will need to take the following steps for a workaround:
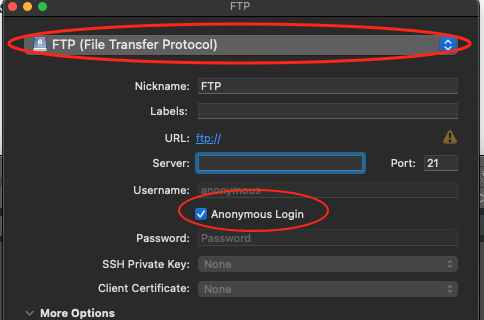
Result:
Using the Downloader Script for S3 (To be updated soon) ¶
The Python script located here allows you to specify a number of options so to download a customized subset of the sample from the S3 bucket. In particular, it allows you to specify:
- Age Ranges
- Sex
- Handedness
- Study and Visit
- Scan Type
- Series
If none of these items are specified, the script will assume that you would like to download all available raw NKI-RS Neuroimaging Data release data.
The script requires the following dependencies to function properly:
- Python version 2.7 or 3.6
- pandas
- boto3
To install pandas and boto3 within Python, you can use the following command: pip install pandas boto3
Note: All examples in this section assume some literacy with the Unix command line and that the commands are being executed from the local directory where the script is stored. If you have not used the command line before, a number of tutorials to get you started are linked to here.
Study / Visit Codes¶
If you want to specify which studies / visits to download, you must take a study code and append the desired visit code to it, and then add this after the -v option. You may specify multiple study/visit code combinations (using the NITRC/AWS convention) after this option.
For example:
python download_rockland_raw_bids_ver2.py -o /data/output -v BAS1
This will download all of the files for the Baseline visit and store them in /data/output in BIDS format.
Scan Types¶
Multiple scan types, separated by spaces after the scan type parameter, can be selected. These scan types can take on the following values, which come from the BIDS specification:
- anat - Anatomical scans.
- func - Functional/resting-state scans.
- dwi - DTI scans.
For example:
python download_rockland_raw_bids_ver2.py -o /data/output -t anat dwi
This will download all of the anatomical and dti data only across all visits.
Series Codes¶
Similarly, multiple series can be specified at the command line. These series can take on the following values:
- Non-neurofeedback studies:
- REST645
- REST1400
- RESTCAP
- RESTPCASL
- CHECKERBOARD645
- BREATHHOLD1400
- CHECKERBOARD1400
For example:
python download_rockland_raw_bids_ver2.py -o /data/output -e REST645 CHECKERBOARD645
This will download only the data for these two series across all participants and visits.
An Elaborate Example¶
Here is a more complex example of the downloader script’s syntax, which illustrates how you can combine command line options to generate extremely nuanced subsets of the Rockland sample.
python download_rockland_raw_bids_ver2.py -o /data/output -e REST645 -v BAS1 -t anat func -gt 15 -lt 40 -m L -x F
This command would download all anatomical series and all resting state series with a 645 ms TR for left-handed women between the ages of 15 and 40, but only from the abridged and second visits of the discovery science and neurofeedback studies. This data could be used to assess within-subject variations in connectivity and neuroanatomy for this specific population, since the first visit for the discovery science and neurofeedback protocols are identical.
To get more information on all of the parameters you may use, type:
python download_rockland_raw_bids_ver2.py -h
(No longer supported and/or updated ) Downloading Neuroimaging Data Releases from NITRC ¶
Anonymized image data is available in either DICOM or NIfTI file formats on NITRC. Each scan folder also includes a JSON file containing information on scan parameters and file provenance (any changes made to the image since it came out of the scanner).
In order to access NKI-RS neuroimaging data via NITRC you must be:
- Registered for NITRC. You may register here if you are not registered already.
- Logged into NITRC.
- Registered with the 1000 Functional Connectomes Project.
Once these conditions are satisified, you may use the links below to download data. If you are not logged in and properly registered a permissions error will occur.
NIfTI Files Downloads
DICOM Files Downloads
- Release 1 (181 subjects)
- Release 2 (14 subjects)
- Release 3 (46 subjects)
- Release 4 (88 Subjects)
- Release 5 (89 Subjects)
- Release 6 (151 Subjects)
- Release 7 (88 Subjects)
- Release 8 (114 Subjects)
Physiological Data (cardiac and respiratory recordings) Downloads
- Release 1 (For initial subjects in Release 1, cardiac data has been excluded due to a technical error; respiratory data was unaffected. Click here for a listing of affected subjects.
- Release 2
- Release 3
Motion Statistics
Release 1
Release 2
Release 3
Release 4
Release 5
Minimal Phenotypic Information (age, sex, handedness) Downloads
- The BIDS participants.tsv file contains minimal phenotypic information for all releases.

